Objectives
BYTE-Sea coordinates the development of interfaces for data access, progress monitoring, and management of the ATLASea programme.
1. Interoperability and Data Publication
The BYTE-Sea project ensures the interoperability and security of data produced by the ATLASea programme, and facilitates its dissemination in line with FAIR principles and Open Science practices. In collaboration with the SEQ-Sea project, it supports the validation and publication of genomic data in reference repositories (such as European Nucleotide Archive – ENA), including raw reads, assembled genomes, and both structural and functional annotations.
2. Data Visualisation
A data portal allows users to track analysed species, sampling events, and the production and publication of genomic data in relation to taxonomic and biodiversity reference systems.
This portal also provides access to genome visualisation tools (e.g., Genome Browser) and similarity search functions, and will soon include first-level analysis tools (for functional and comparative genomics).
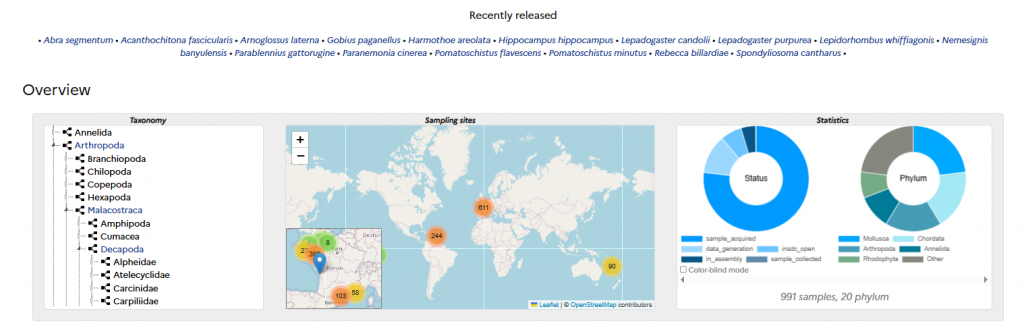
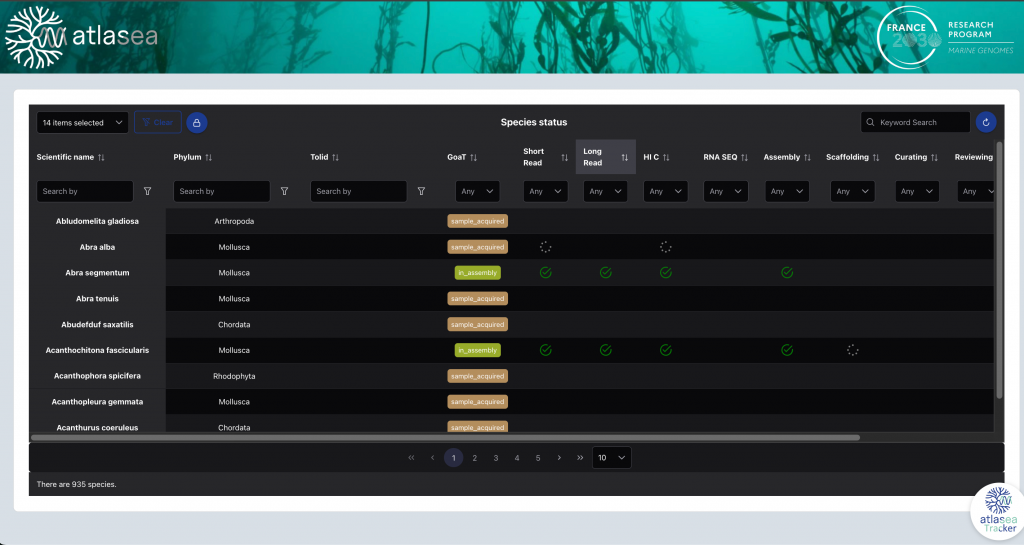
A monitoring portal, currently under development and restricted to consortium members, will enable tracking of sample processing—from taxonomic validation through DNA preparation and sequencing, to the publication of assemblies and annotations in public repositories.
3. Annotation
Predicting gene structure and function in eukaryotes is a complex task that largely depends on existing knowledge and available resources for the species in question. To meet this challenge, the ATLASea programme—which aims to sequence genomes from a wide range of under-studied species with few or no close relatives already sequenced—has adopted a systematic approach.
In collaboration with the SEQ-Sea project, this approach initially involves automating structural and functional annotation pipelines. Subsequently, a community annotation environment will be established to refine gene structure and function predictions following their initial release.
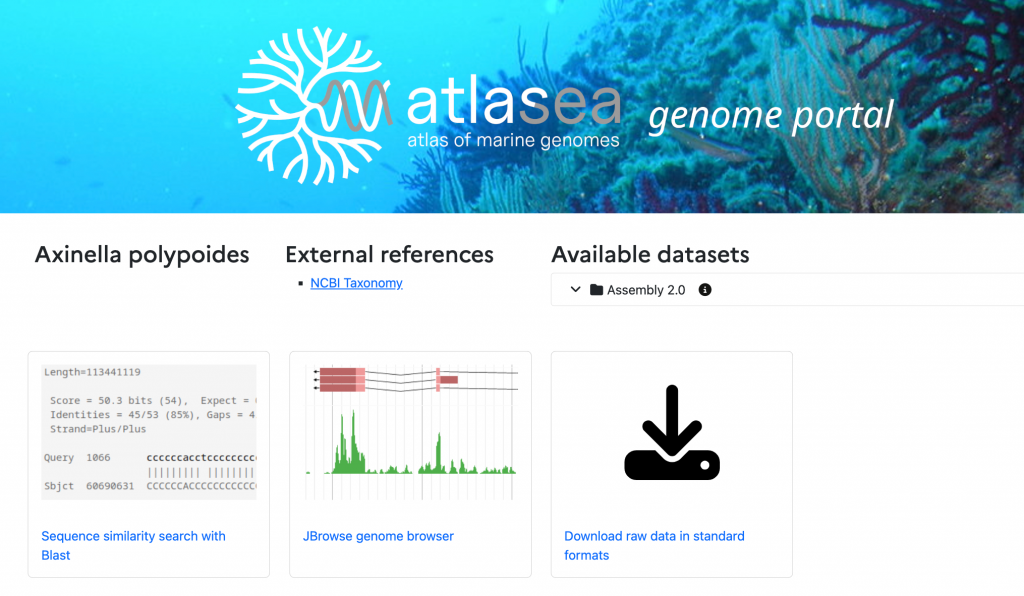
4. Comparative analysis
A dedicated portal is being developed to provide access to all genomic data on marine organisms generated by the ATLASea programme and other consortia. This environment will offer access to high-quality genome assemblies (including chromosome-level sequences).
Available data will include genomic sequences, gene annotations, coding and non-coding regions, and functional annotations. The portal will also offer tools for sequence search, evolutionary and functional analysis, and data visualisation in environmental and biological contexts.
Resources
The project’s main digital infrastructure is hosted at the IFBcore facility (IDRIS-Orsay) and within the BigOuest platform, a federation of the ABiMS (Roscoff) and GenOuest (Rennes) infrastructures.
We rely on storage and computing resources at these data centres, ensuring high data and portal availability by distributing access across three locations: Orsay, Rennes, and Plouzané.
Organisation
Lead organisation : Institut Français de Bioinformatique (IFB)
Manager : Erwan Corre
| Station Biologique de Roscoff Team | Institut de Biologie de l’École Normale Supérieure (IBENS) Team | Institut de Recherche en Informatique et Systèmes Aléatoires (IRISA) Team | Institut Français de Bioinformatique (IFB) Team | Ifremer Team |
| Loraine Guegen Mark Hoebeke Annie Lebreton Alexandre Nicaise Gildas Le Corguille | Lucile Jeusset Samuel Lalam Alexandra Louis Hugues Roest Crollius | Anthony Bretaudeau Théo Foulquier Romane Libouban Mateo Boudet | Nicole Charrière Julien Seiler | Pauline Auffret Patrick Durand Laura Leroi Yaëlle Pihan |
Funding
The exploratory research programme “ATLASea: Atlas of marine genomes” and BYTE-Sea targeted project (ANR-22-EXAT-0004) receive government funding managed by the Agence Nationale de la Recherche (ANR) under the future investment plan “France 2030”.
